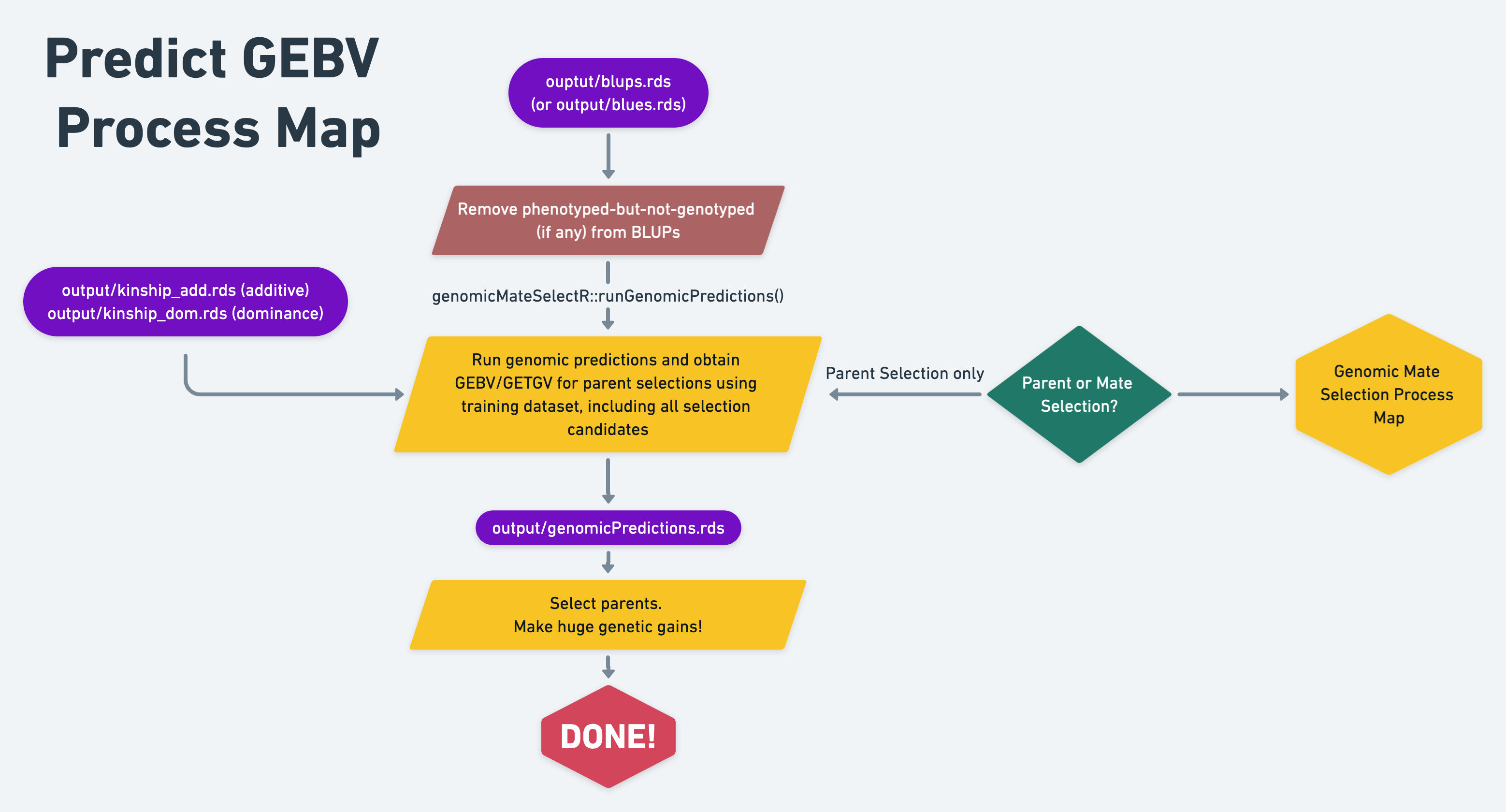
12 Predict parental breeding values
Now that we tested genomic prediction accuracy using cross-validation, we can run genomic predictions.
In the previous section where we introduced genomic prediction, we learned how to use the mmer() function in library(sommer) to run GBLUP models and also rrBLUP models.
For the actual predictions, we can use the function build into library(genomicMateSelectR), runGenomicPredictions(). You can find the documentation for that function by clicking here.
runGenomicPredictions() is a wrapper that uses mmer() under-the-hood. It expects de-regressed BLUPs and weights as input.
12.2 Set-up for the predictions
Similar set-up to what we did for cross-validation.
Load the BLUps and the kinship matrix.
blups<-readRDS(here::here("output","blups.rds"))
A<-readRDS(file=here::here("output","kinship_add.rds"))
blups %<>%
# based on cross-validation, decided to exclude MCMDS from this analysis
filter(Trait != "MCMDS") %>%
# need to rename the "blups" list to comply with the runCrossVal function
rename(TrainingData=blups) %>%
dplyr::select(Trait,TrainingData) %>%
# need also to remove phenotyped-but-not-genotyped lines
mutate(TrainingData=map(TrainingData,
~filter(.,germplasmName %in% rownames(A)) %>%
# rename the germplasmName column to GID
rename(GID=germplasmName)))
blups
#> # A tibble: 3 × 2
#> Trait TrainingData
#> <chr> <list>
#> 1 DM <tibble [346 × 6]>
#> 2 logFYLD <tibble [350 × 6]>
#> 3 logDYLD <tibble [348 × 6]>Selection index:
SIwts<-c(DM=15,
#MCMDS=-10,
logFYLD=20,
logDYLD=20)
SIwts
#> DM logFYLD logDYLD
#> 15 20 20Only difference: do not subset the kinship matrix. Or more precisely, keep any genotypes meant to be either in the training set (phenotyped-and-genotyped) and those that are selection candidates (not-necessarily-genotyped).
In this example, simply leave all lines in the kinship matrix.
12.3 Run genomic predictions
gpreds<-runGenomicPredictions(modelType="A",
selInd=TRUE, SIwts=SIwts,
blups=blups,
grms=list(A=A),
ncores=3)
#> Loading required package: furrr
#> Loading required package: future
#> iteration LogLik wall cpu(sec) restrained
#> 1 -187.325 9:29:57 0 0
#> 2 -187.167 9:29:57 0 0
#> 3 -187.095 9:29:57 0 0
#> 4 -187.077 9:29:58 1 0
#> 5 -187.075 9:29:58 1 0
#> 6 -187.075 9:29:58 1 0
#> iteration LogLik wall cpu(sec) restrained
#> 1 -135.425 9:29:58 1 0
#> 2 -135.406 9:29:58 1 0
#> 3 -135.395 9:29:58 1 0
#> 4 -135.391 9:29:58 1 0
#> 5 -135.39 9:29:58 1 0
#> iteration LogLik wall cpu(sec) restrained
#> 1 -146.037 9:29:58 0 0
#> 2 -146.031 9:29:58 0 0
#> 3 -146.028 9:29:58 0 0
#> 4 -146.027 9:29:59 1 012.4 Extract GEBV
Let’s look at the output.
gpreds
#> # A tibble: 1 × 2
#> gblups genomicPredOut
#> <list> <list>
#> 1 <tibble [963 × 6]> <tibble [3 × 4]>We have a single-row tibble.
To access a simple table listing GEBV for each trait and the selection index:
gpreds$gblups[[1]]
#> # A tibble: 963 × 6
#> GID predOf SELIND DM logFYLD logDYLD
#> <chr> <chr> <dbl> <dbl> <dbl> <dbl>
#> 1 IITA-TMS-IBA30572 GEBV -7.85 -0.610 0.0303 0.0348
#> 2 IITA-TMS-IBA940237 GEBV 1.04 0.0983 -0.0124 -0.00961
#> 3 IITA-TMS-IBA961642 GEBV 15.0 0.808 0.0741 0.0713
#> 4 IITA-TMS-ONN920168 GEBV 3.65 0.284 -0.0129 -0.0172
#> 5 IITA-TMS-WAR4080 GEBV -6.53 -0.529 0.0313 0.0391
#> 6 IITA-TMS-WAR4092 GEBV -6.28 -0.513 0.0305 0.0404
#> 7 IITA-TMS-WAR820249 GEBV 3.33 0.0289 0.0752 0.0699
#> 8 IITA-TMS-WAR820422 GEBV 4.11 0.0639 0.0831 0.0746
#> 9 IITA-TMS-WAR940009 GEBV 13.2 0.749 0.0482 0.0480
#> 10 IITA-TMS-WAR940017 GEBV -15.3 -1.12 0.0463 0.0240
#> # … with 953 more rowsAt this point, you can use the SELIND predictions directly to rank and select parents.
Example: sort by SELIND and pick the top 10…
gpreds$gblups[[1]] %>%
arrange(desc(SELIND)) %>%
slice(1:10)
#> # A tibble: 10 × 6
#> GID predOf SELIND DM logFYLD logDYLD
#> <chr> <chr> <dbl> <dbl> <dbl> <dbl>
#> 1 TMS13F1307P0008 GEBV 26.7 1.59 0.0540 0.0880
#> 2 TMS14F1035P0004 GEBV 26.5 1.63 0.0264 0.0774
#> 3 TMS14F1262P0002 GEBV 24.3 1.35 0.0877 0.110
#> 4 TMS19F1091P0065 GEBV 22.4 1.25 0.0742 0.105
#> 5 TMS14F1303P0012 GEBV 22.2 1.44 0.00866 0.0197
#> 6 TMS14F1312P0003 GEBV 22.0 1.38 0.0258 0.0413
#> 7 TMS19F1041P0112 GEBV 20.5 1.05 0.104 0.134
#> 8 TMS19F1050P0056 GEBV 20.3 1.25 0.0175 0.0615
#> 9 TMS14F1284P0019 GEBV 20.2 1.39 -0.0310 -0.00172
#> 10 IITA-TMS-ZAR000120 GEBV 19.9 1.32 -0.0156 0.0209For more detailed output, including variance component estimates:
gpreds$genomicPredOut[[1]]
#> # A tibble: 3 × 4
#> Trait gblups varcomps fixeffs
#> <chr> <list> <list> <list>
#> 1 DM <tibble [963 × 2]> <df [2 × 4]> <df [1 × 5]>
#> 2 logFYLD <tibble [963 × 2]> <df [2 × 4]> <df [1 × 5]>
#> 3 logDYLD <tibble [963 × 2]> <df [2 × 4]> <df [1 × 5]>
gpreds$genomicPredOut[[1]]$varcomps[[1]]
#> VarComp VarCompSE Zratio
#> u:GIDa.drgBLUP-drgBLUP 1.427019 0.5677452 2.513485
#> units.drgBLUP-drgBLUP 4.766819 0.4955154 9.619921
#> Constraint
#> u:GIDa.drgBLUP-drgBLUP Positive
#> units.drgBLUP-drgBLUP Positive